Brain tumor
Brain Tumor is a very rare
condition that very few people usually affected by it. Of any aging brain,
tumors can occur, It usually occurs in fifth or sixth decades of the patients
i.e., after fifty years. To improve human judgement in diagnosis advent of new technology into health care can be witnessed. An entrance of computer vision into diagnosis would reduce human error in judgment. The MRI scans are capable of identifying even the smallest aberrations in the human body.
Brain Tumor Types
In the brain, you can get two
types of tumors. They are
1) Benign Tumors:
Benign tumor is completely
removed from the brain, can be completely removed. Usually, do not have any
problem in their future life.
2) Malignant Tumors:
Malignant tumor can be removed or it can
cause recurrence of the brain, but not spread outside of the brain. It recurs
into the brain.
Brain Tumor Diagnosis –
Diagnosis of Brain Tumor can be
done by either doing the CT scan of the brain or MRI of the Brain. MRI usually
cannot miss any brain tumor, CT scan sometimes misses brain tumors.
Deep learning
Here we train a model to specifically identify these tiny aberrations from MRIs and predict presence of a tumor with high accuracy. Convolutional Neural Network (CNN) is one of the most effective techniques for this problem statement. Thus using image preprocessing and deep learning using keras and tensorflow, we built a highly reliant and robust model to solve this problem.
The source code for Convolutional Neural Network is
Initially the modeules such as tensorflow, keras, numpy and matplotlib are imported.
- import tensorflow as tf
- import numpy as np
- from keras.preprocessing import image
- from keras.preprocessing.image import ImageDataGenerator
- from matplotlib import pyplot as plt
- tf.__version__
Then Data Preprocessing is done with the image training dataset
- train_datagen = ImageDataGenerator(rescale = 1./255, shear_range = 0.2, zoom_range = 0.2, horizontal_flip = True)
- training_set = train_datagen.flow_from_directory('dataset/training_set', target_size = (64, 64), batch_size = 32, class_mode = 'binary')
Alternatively the preprocessing is done with the image test dataset
- test_datagen = ImageDataGenerator(rescale = 1./255)
- test_set = test_datagen.flow_from_directory('dataset/test_set', target_size = (64, 64), batch_size = 32, class_mode = 'binary')
Followed by training and testing the CNN is build with differnt cnn layers such as the convolution layer, maxpool layer, flatten layer, connection layer and the output layer.
- cnn = tf.keras.models.Sequential()
- cnn.add(tf.keras.layers.Conv2D(filters=32, kernel_size=3, activation='relu', input_shape=[64, 64, 3]))
- cnn.add(tf.keras.layers.MaxPool2D(pool_size=2, strides=2))
- cnn.add(tf.keras.layers.Conv2D(filters=64, kernel_size=3, activation='relu'))
- cnn.add(tf.keras.layers.MaxPool2D(pool_size=2, strides=2))
- cnn.add(tf.keras.layers.Flatten())
- cnn.add(tf.keras.layers.Dense(units=128, activation='relu'))
- cnn.add(tf.keras.layers.Dense(units=1, activation='sigmoid'))
Once all the layers of CNN are added training is done. For training the optimizer used is 'adam' and 'accuracy' is used as an metrics.
- cnn.compile(optimizer = 'adam', loss = 'binary_crossentropy', metrics = ['accuracy'])
- cnn.fit(x = training_set, validation_data = test_set, epochs = 15)
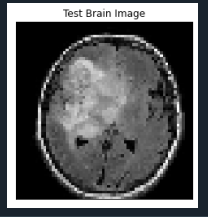
Finally after training CNN, testing is done with an brain tumor image. This final testing code is meant to predict whether the given MRI image is affected y brain tumor or not.
- test_image = image.load_img('dataset/2.jpg', target_size = (64, 64))
- plt.imshow(test_image)
- plt.title('Test Brain Image'), plt.xticks([]), plt.yticks([])
- plt.show()
- test_image = image.img_to_array(test_image)
- test_image = np.expand_dims(test_image, axis = 0)
- result = cnn.predict(test_image)
- training_set.class_indices
- if result[0][0] == 1:
- prediction = 'brain tumor present'
- else:
- prediction = 'no brain tumor'
- print(prediction)
The output for this code is:
Input test image
Predicted Image
The demo of this code is given in the following link:
For queries contact : researchscholarguidance@gmail.com